Publications
Filter by type:
Sparse MoEs meet Efficient Ensembles
Machine learning models based on the aggregated outputs of submodels, either at the activation or prediction levels, lead to strong performance. We study the interplay of two popular classes of such models: ensembles of neural networks and sparse mixture of experts (sparse MoEs). First, we show that these two approaches have complementary features whose combination is beneficial. Then, we present partitioned batch ensembles, an efficient ensemble of sparse MoEs that takes the best of both classes of models. Extensive experiments on fine-tuned vision transformers demonstrate the accuracy, log-likelihood, few-shot learning, robustness, and uncertainty calibration improvements of our approach over several challenging baselines. Partitioned batch ensembles not only scale to models with up to 2.7B parameters, but also provide larger performance gains for larger models.
Plex: Towards Reliability using Pretrained Large Model Extensions
A recent trend in artificial intelligence is the use of pretrained models for language and vision tasks, which have achieved extraordinary performance but also puzzling failures. Probing these models’ abilities in diverse ways is therefore critical to the field. In this paper, we explore the reliability of models, where we define a reliable model as one that not only achieves strong predictive performance but also performs well consistently over many decision-making tasks involving uncertainty (e.g., selective prediction, open set recognition), robust generalization (e.g., accuracy and proper scoring rules such as log-likelihood on in- and out-of-distribution datasets), and adaptation (e.g., active learning, few-shot uncertainty). We devise 10 types of tasks over 40 datasets in order to evaluate different aspects of reliability on both vision and language domains. To improve reliability, we developed ViT-Plex and T5-Plex, pretrained large model extensions for vision and language modalities, respectively. Plex greatly improves the state-of-the-art across reliability tasks, and simplifies the traditional protocol as it improves the out-of-the-box performance and does not require designing scores or tuning the model for each task. We demonstrate scaling effects over model sizes up to 1B parameters and pretraining dataset sizes up to 4B examples. We also demonstrate Plex’s capabilities on challenging tasks including zero-shot open set recognition, active learning, and uncertainty in conversational language understanding.
Ensembles of Classifiers: a Bias-Variance Perspective
Ensembles are a straightforward, remarkably effective method for improving the accuracy, calibration, and robustness of neural networks on classification tasks. Yet, the reasons underlying their success remain an active area of research. Building upon (Pfau, 2013), we turn to the bias-variance decomposition of Bregman divergences in order to gain insight into the behavior of ensembles under classification losses. Introducing a dual reparameterization of the bias-variance decomposition, we first derive generalized laws of total expectation and variance, then discuss how bias and variance terms can be estimated empirically. Next, we show that the dual reparameterization naturally introduces a way of constructing ensembles which reduces the variance and leaves the bias unchanged. Conversely, we show that ensembles that directly average model outputs can arbitrarily increase or decrease the bias. Empirically, we see that such ensembles of neural networks may reduce the bias. We conclude with an empirical analysis of ensembles over neural network architecture hyperparameters, revealing that these techniques allow for more efficient bias reduction than standard ensembles.
Faster & More Reliable Tuning of Neural Networks: Bayesian Optimization with Importance Sampling
Many contemporary machine learning models require extensive tuning of hyperparameters to perform well. A variety of methods, such as Bayesian optimization, have been developed to automate and expedite this process. However, tuning remains costly as it typically requires repeatedly fully training models. To address this issue, Bayesian optimization has been extended to use cheap, partially trained models to extrapolate to expensive complete models. This approach enlarges the set of explored hyperparameters, but including many low-fidelity observations adds to the intrinsic randomness of the procedure and makes extrapolation challenging. We propose to accelerate tuning of neural networks in a robust way by taking into account the relative amount of information contributed by each training example. To do so, we leverage importance sampling (IS); this significantly increases the quality of the function evaluations, but also their runtime, and so must be done carefully. Casting hyperparameter search as a multi-task Bayesian optimization problem over both hyperparameters and IS design achieves the best of both worlds. By learning a parameterization of IS that tradeso↵ evaluation complexity and quality, our method improves upon validation error, in the average and worst-case, while using higher fidelity observations with less data. We show that this results in more reliable performance of our method in less wall-clock time across a variety of datasets and neural architectures.
Distilling Ensembles Improves Uncertainty Estimates
We seek to bridge the performance gap between batch ensembles (ensembles of deep networks with shared parameters) and deep ensembles on tasks which require not only predictions, but also uncertainty estimates for these predictions. We obtain negative theoretical results on the possibility of approximating deep ensemble weights by batch ensemble weights, and so turn to distillation. Training a batch ensemble on the outputs of deep ensembles improves accuracy and uncertainty estimates, without requiring hyper-parameter tuning. This result is specific to the choice of batch ensemble architectures: distilling deep ensembles to a single network is unsuccessful, despite single networks having only marginally fewer parameters than batch ensembles.
Population-Based Black-Box Optimization for Biological Sequence Design
The use of black-box optimization for the design of new biological sequences is an emerging research area with potentially revolutionary impact. The cost and latency of wet-lab experiments requires methods that find good sequences in few experimental rounds of large batches of sequences–a setting that off-the-shelf black-box optimization methods are ill-equipped to handle. We find that the performance of existing methods varies drastically across optimization tasks, posing a significant obstacle to real-world applications. To improve robustness, we propose Population-Based Black-Box Optimization (P3BO), which generates batches of sequences by sampling from an ensemble of methods. The number of sequences sampled from any method is proportional to the quality of sequences it previously proposed, allowing P3BO to combine the strengths of individual methods while hedging against their innate brittleness. Adapting the hyper-parameters of each of the methods online using evolutionary optimization further improves performance. Through extensive experiments on in-silico optimization tasks, we show that P3BO outperforms any single method in its population, proposing higher quality sequences as well as more diverse batches. As such, P3BO and Adaptive-P3BO are a crucial step towards deploying ML to real-world sequence design.
Deep uncertainty and the search for proteins
Machine learning applications to molecule and protein design require models that provide meaningful uncertainty estimates. For example, Bayesian optimization for biomolecules searches through the space of viable designs, trading off the exploration of uncertain regions with the exploitation of high-value areas. We introduce protein optimization datasets as a benchmarking environment for ML uncertainty on real-world distribution shifts; investigate scalable models robust to the distribution shift inherent to large-batch, multi-round BO over protein space; and show that intra-ensemble diversification improves calibration on multi-round regression tasks, allowing for more principled biological compound design.
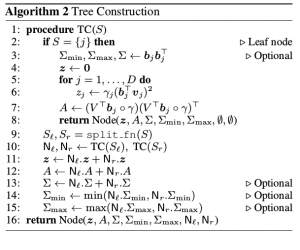
It is often desirable for recommender systems and other information retrieval applications to provide diverse results, and determinantal point processes (DPPs) have become a popular way to capture the trade-off between the quality of individual results and the diversity of the overall set. However, computational concerns limit the usefulness of DPPs in practice. Sampling from a DPP is inherently expensive: if the underlying collection contains N items, then generating each DPP sample requires O(N) time following a one-time preprocessing phase. Additionally, results often need to be personalized to a user, but standard approaches to personalization invalidate the preprocessing, making personalized samples especially expensive. In this work we address both of these shortcomings. First, we propose a new algorithm for generating DPP samples in O(log N) time following a slightly more expensive preprocessing phase. We then extend the algorithm to support arbitrary query-time feature weights, allowing us to generate samples customized to individual users while still retaining logarithmic runtime. Experiments show that our approach runs over 300 times faster than traditional DPP sampling on collections of 100,000 items for samples of size 10.
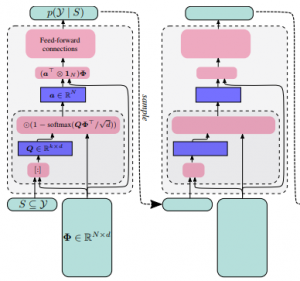
DppNet: Approximating Determinantal Point Processes with Deep Networks
Determinantal Point Processes (DPPs) provide an elegant and versatile way to sample sets of items that balance the point-wise quality with the set-wise diversity of selected items. For this reason, they have gained prominence in many machine learning applications that rely on subset selection. However, sampling from a DPP over a ground set of size N is a costly operation, requiring in general an O(N^3) preprocessing cost and an O(Nk^3) sampling cost for subsets of size k. We approach this problem by introducing DPPNets: generative deep models that produce DPP-like samples for arbitrary ground sets. We develop an inhibitive attention mechanism based on transformer networks that captures a notion of dissimilarity between feature vectors. We show theoretically that such an approximation is sensible as it maintains the guarantees of inhibition or dissimilarity that makes DPPs so powerful and unique. Empirically, we demonstrate that samples from our model receive high likelihood under the more expensive DPP alternative.
Biological Sequences Design using Batched Bayesian Optimization
Being able to effectively design biological sequences like DNA and proteins would have transformative impact on medicine. Currently, the most popular method in the life sciences for performing design is directed evolution, which explores sequence space by making small mutations to existing sequences. Alternatively, Bayesian optimization (BO) provides an attractive framework for model-based black-box optimization, and has achieved many recent successes in life sciences applications. However, within the ML community, most large-scale BO efforts have focused on hyper-parameter tuning. These methods often do not translate to biological sequence design, where the search space is over a discrete alphabet, wet-lab experiments are run with considerable parallelism (1K-100K sequences per batch), and experiments are sufficiently slow and expensive that only few rounds of experiments are feasible. This paper discusses the particularities of batched BO on a large discrete space, and investigates the design choices that must be made in order to obtain robust, scalable, and experimentally successful models within this unique context.
